'Gram negative' bacterial cells such as H. influenzae have two membranes enclosing their cytoplasm, and DNA must travel across both of these and the intervening layer of periplasm and mesh-like cell wall. Here I'll be talking mainly about uptake across the H. influenzae outer membrane.
Transport of DNA across this or any membrane is predicted to be difficult because the membrane is hydrophobic and DNA is hydrophilic. That is, the membrane is made of lipids and repels charged molecules including water, whereas DNA has lots of negative charge and is surrounded by water and positively charged ions. Thus DNA and membranes will repel each other.
Because lots of valuable nutrients are hydrophilic, cells have evolved protein-lined pores in their membranes through which particular charged molecules can pass. But DNA presents
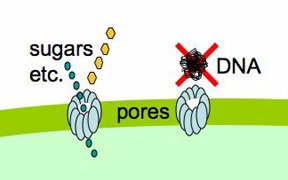 special problems, because it exists as very long stiff fibers, much much longer than the small sugar and amino acid molecules that most pores transport. Nevertheless, cells easily take up DNA molecules more than 50,000 base pairs long, even though these molecules are longer than the cells. A DNA fiber is very thin, and on the scale of a cell it's very flexible, so there's no problem fitting this much DNA into the cell. In fact H. influenzae cells already contain about 40 times this much DNA in their own chromosomes.
special problems, because it exists as very long stiff fibers, much much longer than the small sugar and amino acid molecules that most pores transport. Nevertheless, cells easily take up DNA molecules more than 50,000 base pairs long, even though these molecules are longer than the cells. A DNA fiber is very thin, and on the scale of a cell it's very flexible, so there's no problem fitting this much DNA into the cell. In fact H. influenzae cells already contain about 40 times this much DNA in their own chromosomes.DNA uptake is problematic because, on a molecular scale, DNA is quite stiff. Its stiffness is described by its 'persistence length', which is about 50nm. As typical pores are only a few nm across, this means that DNA won't spontaneously collapse into a clump small enough that can pass through such a pore (like the DNA shown above with a big red X).
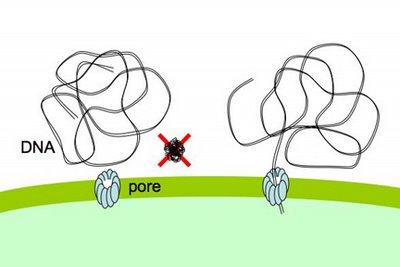
The DNA fiber is only about 2nm across, so its stiffness wouldn't be a problem if it could be 'threaded' through a pore, starting from one end. Work mainly from other labs strongly suggests that the channel is formed by the same proteins that form the channel used by a protein filament, the type 4 pilus, which cells can extend and retract from their surface. These channels are about 6nm across, more than wide enough for DNA to thread through.
But I suspect that the uptake machinery doesn't need to find the DNA's end in order to take it up. That is, I don't think DNA is usually threaded through the pilin channel.
The evidence for end-independent DNA uptake is that competent cells have no trouble transporting circular plasmids across the outer membrane, and that the plasmids remain unbroken in the periplasm (though they can't be transported across the inner membrane without being cut or broken). I need to reevaluate the strength of the evidence that the plasmid DNA doesn't get broken, but for now let's assume that it's solid.
How then does the plasmid get across the membrane? I propose that the DNA is sharply bent or kinked by the uptake machinery, allowing it to be poked in through the pilin channel. I say
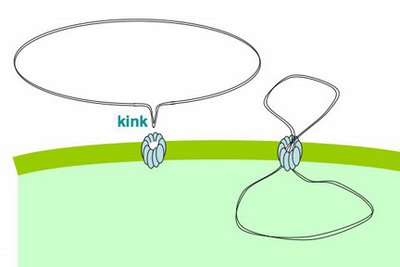 'by the uptake machinery' because normal DNA doesn't spontaneously form kinks, though it can be a bit curved. I also propose that one reason for preferential uptake of DNA with USS sequences is that these sequences are particularly easy to bend or kink. Once the kinked DNA is poked through the channel, the rest of it can be pulled in without additional kinking. This hypothetical process is illustrated in the little movie on our lab home page. The movie also shows the end of the DNA being pulled into the periplasm, and one strand of the DNA then being transported across the inner membrane.
'by the uptake machinery' because normal DNA doesn't spontaneously form kinks, though it can be a bit curved. I also propose that one reason for preferential uptake of DNA with USS sequences is that these sequences are particularly easy to bend or kink. Once the kinked DNA is poked through the channel, the rest of it can be pulled in without additional kinking. This hypothetical process is illustrated in the little movie on our lab home page. The movie also shows the end of the DNA being pulled into the periplasm, and one strand of the DNA then being transported across the inner membrane.Unfortunately, I don't have any direct evidence that DNA with the USS sequence is any easier to kink than other DNA.
One way to create a kink would be to separate the two DNA strands (single strands are much more flexible than a double-helix). The AT-rich sequences on one side of the USS core are good candidates for this kind of kinking, because their weak base pairs are more easily separated than GC base pairs. But I have only a very naive understanding of how this and other forces interact to determine whether a given DNA sequence can easily be kinked. I think it's time to consult an expert.
If the issue was simply that of kinking, shouldn't there be a whole family of sequences that H. influenzae happily takes up?
ReplyDeleteIf that were the case, the over-representation of the 9bp USS motif in H. influenzae's own genome would no longer make sense to me!
The uptake machinery issues may include firmly grabbing hold of and orienting correctly and kinking and pushing, and different parts of the motif may contribute differently to these functions.
ReplyDeleteWhat about if the motif is used as a start to cleave and thread the DNA into the cell ?
ReplyDeleteI also agree with the first comment that if was just there for kinking then there would be a lot more sequences capable of the same structure.