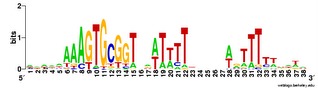
 Not only did the different replicates find different numbers of sites (between 21 and 28), they settled on motifs with slightly different centers and (once) a different length. This gives me more confidence that this consensus logo represents most of the variation in sites.
Not only did the different replicates find different numbers of sites (between 21 and 28), they settled on motifs with slightly different centers and (once) a different length. This gives me more confidence that this consensus logo represents most of the variation in sites.One remaining concern is that these searches were not unbiased, in that I started them all with a 'fragmentation mask' that specified the positions of gaps in the motif. See this post for an explanation. What would a completely unbiased search find? I suspect it will close the gaps, and ignore the consensus at positions that aren't close to the core, as this is what happens in unbiased searches of the genome. But I should try (now).
...Short break while I do this (28 short sequences means it runs very fast)...
Well I'll be hornswoggled! (Sorry, Language Log influence.) It found the reverse version of the full USS motif!
...Another slightly longer break while I do ten replicate runs...
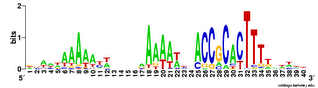 And here's two versions of the logo from the unbiased searches. The top one is what the searches find - the USS motif in reverse orientation . The lower logo is the reverse complement of this (same data, but in back-to-front order and with all the As
And here's two versions of the logo from the unbiased searches. The top one is what the searches find - the USS motif in reverse orientation . The lower logo is the reverse complement of this (same data, but in back-to-front order and with all the As  changed to Ts, Ts to As, Gs to Cs and Cs to Gs).
changed to Ts, Ts to As, Gs to Cs and Cs to Gs).This 'unbiased' motif is actually a better match to the genome consensus than is the motif I got using the fragmentation mask. How nice!
I've been (sometimes) putting 'unbiased' in quotes, because I don't think any pattern search can be truly unbiased. The Gibbs motif sampler program I'm using for these searches has a bias towards compact motifs - there's a built-in penalty for introducing gaps.
No comments:
Post a Comment
Markup Key:
- <b>bold</b> = bold
- <i>italic</i> = italic
- <a href="http://www.fieldofscience.com/">FoS</a> = FoS