
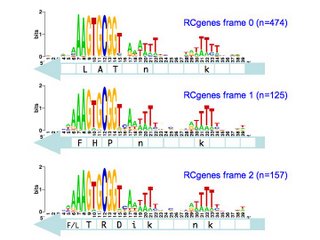
I've finished the reading frame analysis and here are the results. Each logo shows the pattern for the within-gene USSs in a specific position and orientation with respect to the reading frame of the genes they are in. The light-blue arrow below each logo shows the direction in which those USSs' genes are translated into proteins The white boxes superimposed on the arrows show the codons, with the one-letter amino acid code for those amino acids that are more- or less- strongly specified by the USS bases.
So the top left logo summarizes the 49 USSs whose positions and reverse orientation in their genes has their cores specifying the amino acids lysine (K; AAA), valine (V; GTG) and arginine (R; CGG). The consensus of the flanking regions are strong enough that they also are likely to specify particular amino acids, in this case another lysine and a phenylalanine (f; TTT). I put these last two letters in lower case to indicate that the consensus is relatively weak.
So what does this mean? We're still thinking about that. The numbers of sites (n=#, in blue) are sufficiently large that the differences between the logos are significant. I hope we will be able to correlate them to the specific coding constraints, but that's a complex analysis I'm not quite ready for yet.
No comments:
Post a Comment
Markup Key:
- <b>bold</b> = bold
- <i>italic</i> = italic
- <a href="http://www.fieldofscience.com/">FoS</a> = FoS