First, here's a diagram showing the toxTA operon and the mutants we're examining:
The grey bars show the extents of the deletions. The ∆toxT and ∆toxTA mutants have a SpcR/strR cassette inserted at the deletion point, but the ∆toxA mutant has only a short 'scar' sequence at the deletion point.
A few months ago I wrote a post about evidence that ToxA prevents transcription of the toxTA operon from an unexpected internal promoter. Here's a better version of the graph I showed there (note that transcription is going from right to left):
It looks like there are two promoters. The CRP-S promoter is competence-induced so inactive in the M0 samples and maximally active in the M2 samples. The unexpected internal promoter (labeled 'toxTA' in red) is not active in wildtype but highly active in all the ∆toxA samples. Really I should consistently refer to this as the 'putative' internal promoter, but I won't bother for this post.
The line heights in the above graphs are hard to interpret because of the log scale, so here's a linear scale view.
These expression patterns appear to show normal repression of the 'toxTA" internal promoter by ToxA, and release from repression in the ∆toxA mutant. The internal location of the ToxA-repressed promoter is unexpected, but its repression by ToxA fits the pattern observed for the well-studied homologs of this type of toxin/antitoxin system.
In these homologs, transcription of the operon is actually repressed by a toxin-antitoxin complex, not by antitoxin alone. So we would expect to see that repression of the internal toxTA promoter is also released by knockouts of toxT or of both genes. But that's not what we see.
We can't check the effect of a toxT knockout on transcription from the internal promoter, because the internal promoter is deleted in our toxT mutants. But we can check the effect of deleting both toxT and ToxA, because the deletion in our double mutant starts well downstream of the internal promoter.
Surprisingly, transcription from the internal promoter is not increased in the double mutant. The first figure shows coverage for wildtype, ∆toxA and ∆∆toxTA at time point M0. We see that the CRP-S promoter is inactive in all three strains, and the internal promoter is very active only in ∆toxA (purple). It's actually less active in the double knockout (green) than in KW20 (blue).
This figure shows transcription of the same strains at time point M2, when the CRP-S promoter is most active. Now we see transcription from the CRP-S promoter in all strains, although definitely weaker in ∆∆toxTA. We again see strong transcription from the internal promoter in ∆toxA (purple), but very little in ∆∆toxTA.
Could some sort of artefact be responsible for the apparent lack of transcription from the internal promoter in the double knockout? One likely candidate is read-mapping artefacts caused by the presence of toxTA deletions and insertions in the mutant reads but not in the wildtype genome sequence they are being aligned to.
So we could check for these effects, one of the former honours students took a a set of mutant-specific reference sequences for the toxTA region, and separately aligned each set of mutant reads to its corresponding mutant reference sequence, and explained to me how to examine the reads and coverages using the Integrated Genome Viewer (IGV).
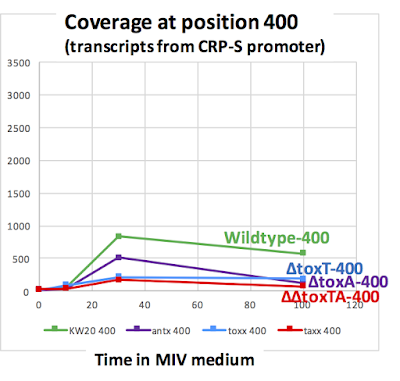
For all the M0 (log phase growth) and M2 samples (max CRP-S induction) I noted the number of reads covering position 400 (at the toxT start codon, ~ 35 bp downstream from the CRP-S promoter, and covering position 500 (about 75 bp downstream from the internal promoter). (I didn't bother analyzing M1 and M3 samples.) I normalized each coverage value by the number of reads in the sample, and calculated the mean coverage over the three replicate samples for each time point.
Here's a linear-scale graph:
And here's the corresponding log-scale graph:
And here's the conclusions:
I'm now going to go back and generate the data for M1 and M3. Then I'll update this post. For now, ignore the notes below.
So, questions we still don't know the answers to:
Why is competence-induced transcription from the CRP-S promoter down modestly in all the toxTA mutants? (Compare actual mean M2 values: wildtype: 830, ∆toxA: 513, ∆toxT: 213; ∆∆toxTA: 173.)
In wildtype cells in log phase, neither antitoxin (ToxA) or or toxin (ToxT) are likely to be present. In wildtype cells at M2, both proteins are likely to be accumulating. We don't know whether there will be more of one than the other - usually the toxin is more stable, and the antitoxin is unstable unless it is bound to toxin. We don't know what the HI0660 toxin's 'toxic' activity is, and we don't know of any other expected activity for the HI0659 antitoxin except binding toxin and repressing toxTA transcription.
The lower transcript levels in the mutants suggests that both ToxA and ToxT contribute positively to transcription from the CRP-S promotor, or to the stability of the resulting transcripts.
Could toxin in wildtype cells, and unopposed toxin in ∆toxA, be somehow stabilizing transcripts? Doesn't make much sense to me.Why is log-phase transcription from the putative internal promoter way up in ∆toxA but way down in ∆∆toxTA? (Compare actual M0 values: wildtype: 134; ∆toxA: 1713; ∆∆toxTA: 170.)
Taken at face value, this would seem to mean that ToxT actively stimulates toxTA transcription, or stabilizes another transcription factor.I hope someone else has some ideas!