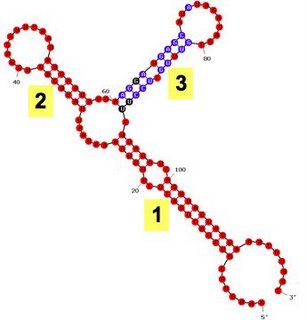
I've been checking the predictions that the RNA-folding program mfold makes about our sxy mRNAs, this time using a feature that colours each position according to how 'well-determined' its pairing assignment is, as shown in the first figure. Here for example the unpaired nucleotide at position 20 is coloured red, meaning that Mfold is quite confident that it is unpaired (a "P-num" score of 0 or 1). Similarly, the paired nucleotide at position 21 is also coloured red to indicate that Mfold is quite confident that it is paired. Many of the nucleotides in stem 3 (positions 61-91) are coloured blue or black, to indicate less confidence (P-num scores of 2 and 3 respectively).
 It's important to remember that the colour doesn't indicate how strong the pairing is, but how good the evidence is (either for or against pairing).
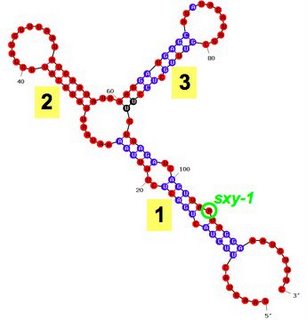
It's important to remember that the colour doesn't indicate how strong the pairing is, but how good the evidence is (either for or against pairing).The second folded molecule is the mutant sxy-1 RNA. The little one-base bubble at the bottom of stem 1 is caused by the mutation. More notable is that most of the nucleotides in stem 1 are now coloured blue rather than red. This means that changing one base from a G to an A has decreased Mfold's confidence in all the base paired positions in the stem. This is consistent with what our RNase digestions show - the one-base sxy-1 change decreases the proportion of molecules that are base paired at all the positions in stem 1, not just at the site of the mutation.
















I don't understand how Mfold determines the strength of pairing, or how to color it I guess. Is there a simple explanation?
ReplyDeleteSimple to say, not simple to do. It calculates the interatomic forces between all (well, maybe not ALL) the parts of the molecule.
ReplyDelete