The folding program Mfold presents only one structure (the first one below) when it folds the sxy mRNA regulatory region (the 50 non-coding bases upstream of the start codon and the first 60 bases of the coding region). I guess this is because other possible foldings are predicted to be much less stable. To get insight into alternative foldings I tried changing the input sequence by making it shorter or longer at the 5' or 3' end. Changing the 5' end might reflect consequences of using different transcription start sites or of binding of a ribosome to its site in front of the start codon. Changing the 3' end might mimic the effect of RNA polymerase stalling or progressing quickly along the DNA. The length changes might also just reveal whether interactions between particular parts of the RNA were sensitive to the presence of other potentially interacting parts.
 Result: There are indeed no alternative stable foldings for RNAs that differ only in small ways from the known regulatory region. However if the RNA is shifted more substantially to include only coding sequences (i.e. the non-coding part of the RNA is not included, and the coding part is extended), another stable folding emerges. This could be biologically significant, if its ability to compete with the 'standard' folding depends on either ribosome binding or rate or transcription. But the factors involved are so complex that I'm starting to see more 'mutant hunts' as our best approach. I'll write about them in a separate post.
Result: There are indeed no alternative stable foldings for RNAs that differ only in small ways from the known regulatory region. However if the RNA is shifted more substantially to include only coding sequences (i.e. the non-coding part of the RNA is not included, and the coding part is extended), another stable folding emerges. This could be biologically significant, if its ability to compete with the 'standard' folding depends on either ribosome binding or rate or transcription. But the factors involved are so complex that I'm starting to see more 'mutant hunts' as our best approach. I'll write about them in a separate post.Thanks to the genome-sequencing efforts of colleagues, we also have sxy sequences from four other (independent) isolates of H. influenzae. Three of these are identical to our KW20 strain, so of course the folding software treats them identically, but the fourth differs at about half a
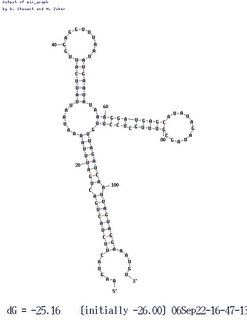
 dozen positions. Three of these differences are in the regulatory region, and they cause an interesting change in folding (shown here). Two of the differences eliminate base pairs in the 'standard' folding, and a third is in an unpaired loop. The new folding puts two of these changed bases into new base pairs, and this folded RNA is predicted to be slightly MORE thermodynamically stable than the standard folding (deltaG = 25.5 rather than 23.1).
dozen positions. Three of these differences are in the regulatory region, and they cause an interesting change in folding (shown here). Two of the differences eliminate base pairs in the 'standard' folding, and a third is in an unpaired loop. The new folding puts two of these changed bases into new base pairs, and this folded RNA is predicted to be slightly MORE thermodynamically stable than the standard folding (deltaG = 25.5 rather than 23.1).In the variant folding the critical stem identified by our hypercompetence mutations is unchanged, but the big loop of RNA that includes the ribosome-binding site and the start codon is folded very differently. However the folding still blocks access of the ribosome to its site, although in a different way. This may be telling us that blocking the ribosome is an important function of the folding. That is, it may be part of the mechanism by which folding of the mRNA regulates its stability or its ability to be translated into protein.
















No comments:
Post a Comment
Markup Key:
- <b>bold</b> = bold
- <i>italic</i> = italic
- <a href="http://www.fieldofscience.com/">FoS</a> = FoS