What works: First, it usually runs without quitting prematurely. Second, it produces what I think is the correct output. I say I think because I don't understand the statistical parts of the output. Third, this time I asked it to search the sequence for 2 motifs rather than 1, and even that seems to be working. Fourth, much of the
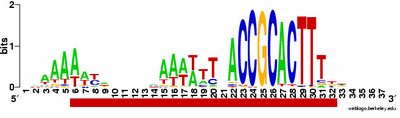 time the output shows the pattern I was expecting to see: alignment of hundreds of short sequences, each containing a sequence related to the previously characterized USS. I trim these down (using Word's search-and-destroy function) and paste them into WebLogo, which generates logos like that above, summarizing the pattern. And it's fast - analyzing the whole genome takes only 5-10 minutes.
time the output shows the pattern I was expecting to see: alignment of hundreds of short sequences, each containing a sequence related to the previously characterized USS. I trim these down (using Word's search-and-destroy function) and paste them into WebLogo, which generates logos like that above, summarizing the pattern. And it's fast - analyzing the whole genome takes only 5-10 minutes.What isn't yet right: Sometimes it misses what should be the very significant motif and instead returns
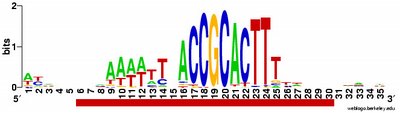 a weak motif that has nothing to do with the USS; I think this means I've set the stringency too low by telling it to expect too many sites with the motif. Often it returns only part of the USS motif, by cutting one side of the USS off, in favour of positions that show no evident similarity at all (when represented as WebLogos). This happens partly because it has decided not to fragment the motif into sub-motifs separated by non-consensus bases - I don't know why. The logo in this paragraph shows such a case. Compare it to the logo in the previous paragraph, and you see that the leftmost AT-rich part is missing. In both images the red underlining shows the positions that the motif search program decided had significant consensuses; in both the program has included positions with no consensuses and left out positions further to the left that would have strong consensuses. It could have included these positions by fragmenting the motif, but it didn't.
a weak motif that has nothing to do with the USS; I think this means I've set the stringency too low by telling it to expect too many sites with the motif. Often it returns only part of the USS motif, by cutting one side of the USS off, in favour of positions that show no evident similarity at all (when represented as WebLogos). This happens partly because it has decided not to fragment the motif into sub-motifs separated by non-consensus bases - I don't know why. The logo in this paragraph shows such a case. Compare it to the logo in the previous paragraph, and you see that the leftmost AT-rich part is missing. In both images the red underlining shows the positions that the motif search program decided had significant consensuses; in both the program has included positions with no consensuses and left out positions further to the left that would have strong consensuses. It could have included these positions by fragmenting the motif, but it didn't.The biggest problem is the mysterious segmentation fault error. If it's using the full genome sequence (1.83megabases), and if I ask it to find a motif bigger than 18bp, the program begins the analysis but stops after a few cycles, reporting a segmentation fault. Googling segmentation fault tells me that this is probably because some string has become too long (the program is trying to put too much information into some location). I'm going to have to read the all-too-terse instructions to see if I can find a way around this. If I can't, I'm hoping that the person who sent me the binary code will take pity on my ignorance and help me solve the problem. The worst case will be if there is no way around this, but even then I think I can still get the analysis I need - it will just take more work on my part, combining results from different parts of the genome.
















No comments:
Post a Comment
Markup Key:
- <b>bold</b> = bold
- <i>italic</i> = italic
- <a href="http://www.fieldofscience.com/">FoS</a> = FoS