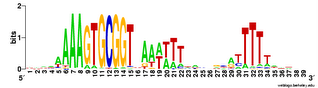
 In addition to the just-posted analysis of genes in coding segments of the genome (the genes), I've analyzed the USSs in the non-coding ('intergenic') segments. Here's the logo.
In addition to the just-posted analysis of genes in coding segments of the genome (the genes), I've analyzed the USSs in the non-coding ('intergenic') segments. Here's the logo.It's a bit different than the in-gene logos in the previous post, and than the whole-genome (genes + intergenic) logo below. At first I was thinking "Great! This logo represents the unconstrained USS, because these USSs don't have to code for protein and so evolve only because of the biased DNA uptake machinery.
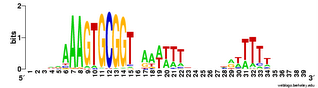
But then I realized I was forgetting about the other function that some USSs appear to have, as molecular signals marking the ends of genes. These 'stop here' signals (called transcription terminators) usually are formed by a palindromic sequence whose RNA can form a GC-rich stem-loop, followed by a short run of Ts (here's an old post that explains palindromes). USSs can do this because two USSs in opposite orientations form a potential palindrome. USSs are over-represented in non-coding regions, and they are particularly common as oppositely-oriented pairs just past the 3' ends of coding regions. RNA transcribed from one of these USSs pairs can form a GC-rich stem, and if the downstream USS is in the forward orientation this will be followed by a run of Ts. Thus these USS pairs are thought to function as transcription terminators.
Only a small subset of all USSs are in pairs that can do this, but they're all in the intergenic regions, and natural selection for the ability to terminate efficiently may constrain the effects of uptake bias. So I really ought to remove the potential terminator USSs from my intergenic dataset before I claim that it represents the unconstrained effects of uptake bias.
This is easier said than done. My initial 'clever' strategy was foiled when I realized I'd made inconsistent changes to the forward and reverse-complement versions of the intergenic sequence set. These changes didn't affect the motif search at all, but they mean that I can't use the motif-search results I have to find pairs of oppositely-oriented USSs. So I queue'd up yet more Gibbs searches, this time searching a single sequence set in both directions. These should show me where the close pairs of USSs are. Then I found a great web site that does all sorts of useful analyses for repeats. (Well, I 'found' it because a new paper from another lab, analyzing USSs in a related species, describes using this site.) The site is great, but sorting out the strands and the directions made my head spin, so I'm waiting for the Gibbs searches to finish before I decide where to apply my limited brain power.
















No comments:
Post a Comment
Markup Key:
- <b>bold</b> = bold
- <i>italic</i> = italic
- <a href="http://www.fieldofscience.com/">FoS</a> = FoS