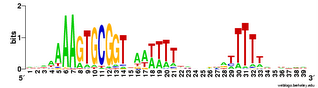
But the Gibbs searches would sometimes run correctly if I only asked them to test 1 or 2 or 3 seeds, so I got some useful data. Here are the results. This logo shows the pattern for the 490 USSs that are neither in coding sequences nor in positions where they are likely to function as transcriptional terminators. So this represents those USSs that are least functionally constrained.
For comparison, here is the logo for all the USSs Gibbs finds in the genome (2136). You can see that the initial As and final Ts are stronger (larger) in the least-constrained USSs. This also makes the USS pattern more strongly
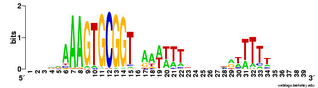 palindromic, so it is symmetric when both DNA strands are considered. To me this suggests that the DNA may kink in the middle, between positions 19 and 20, with base-pair interactions between the initial As and terminal Ts. Tomorrow morning I'm meeting with a structural biochemist who probably set me straight about this. Her main expertise is in protein structure, but at least she'll be able to point me to the best sources of information about DNA kinking.
palindromic, so it is symmetric when both DNA strands are considered. To me this suggests that the DNA may kink in the middle, between positions 19 and 20, with base-pair interactions between the initial As and terminal Ts. Tomorrow morning I'm meeting with a structural biochemist who probably set me straight about this. Her main expertise is in protein structure, but at least she'll be able to point me to the best sources of information about DNA kinking.And here is the complementary logo, based on only the 223 USSs that are both in non-coding regions and in oppositely-oriented pairs close enough to together act as a transcription terminator. The initial As and terminal Ts are still very strong, but now we see a new ACCGCAC pattern on the right, capable of base pairing with the GTGCGGT bases on the left.
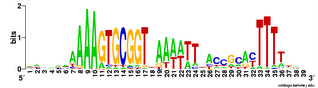 I'm going to have to think more about what this means, as I can't just say "It reflects functional constraints...". (My thinking will mostly consist of drawing sketches on the whiteboards in the hall outside my office.)
I'm going to have to think more about what this means, as I can't just say "It reflects functional constraints...". (My thinking will mostly consist of drawing sketches on the whiteboards in the hall outside my office.)
















For the last logo, the one reflecting transcription terminators....
ReplyDeleteI don't understand why you ask what it means and that you can't just say functional constraints. Don't you predict that if they act as terminators that they would have this stem-loop structure? I am clearly missing something.