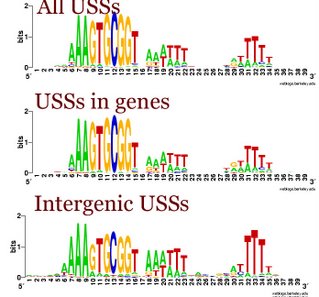
 I had been thinking that the differences were due as much to the terminator function of many intergenic USS as to the coding constraints on the USSs in genes. But when we looked at the logos for the terminator and non-terminator classes of intergenic USSs, we saw that they were quite similar. Compared to the whole-genome USS logo, both showed the strong AAA at the beginning, and strong flanking motifs, especially the TTT at the end. In both the GTGCGGT of the core was also weaker. These patterns are also seen in the logo of all the intergenic USSs to the left.
I had been thinking that the differences were due as much to the terminator function of many intergenic USS as to the coding constraints on the USSs in genes. But when we looked at the logos for the terminator and non-terminator classes of intergenic USSs, we saw that they were quite similar. Compared to the whole-genome USS logo, both showed the strong AAA at the beginning, and strong flanking motifs, especially the TTT at the end. In both the GTGCGGT of the core was also weaker. These patterns are also seen in the logo of all the intergenic USSs to the left.The USSs-in-genes logo (the sum of all 6 reading-frame specific logos I posted a week or so ago) is much more similar to the whole genome logo. This is perhaps not surprising, because most USSs are in genes.
But the similarities of the non-terminator and terminator USSs to each other means that the importance of the initial As and final Ts isn't due to selection for forming a terminator, but to the function of the sequence in DNA uptake. (On reading back over what I posted last week, I see I was pretty much thinking the same thing then. But somehow between then and this morning I got led astray.)
















Isn't the AT content in intergenic regions higher than in genic? Maybe the stronger AT regions you see in the USS are just due to differences in base composition.
ReplyDelete