We do expect that each run of the simulation will come to an equilibrium where the processes enriching the genome for USS are balanced by the processes removing USS from the genome. The difficulty comes in deciding how we will recognize that the simulation has reached this state. Because these are 'stochastic' simulations, the numbers of USS undergo a lot of random fluctuation even at equilibrium, and we need to decide when the long-term average has become sufficiently stable to qualify as 'equilibrium'. When the simulation first starts, the number of USSs increases quickly, but as it gets closer to equilibrium the rate of increase gets slower and slower.
How do we decide when to stop? As with many other things, how we decide to do this will depend on how long we're willing to wait, and on how accurate we want our answers to be.
Some versions of the simulations run fast; these are ones we've set to use small 'genomes' (e.g. 10,000bp) and high mutation rates and high uptake probabilities and (probably strong biases. We can easily let these run for many cycles beyond what looks like equilibrium, so that we're sure that the long-term average USS number is stable despite the short-term fluctuations. I think we should do lots of these, so we have a very clear expectation of what to expect.
But the more realistic versions of the simulations will run much slower, as they'll have larger genomes (maybe even as big as the H. influenzae genome, ~1,830,000bp) and lower mutation rates. These will be run remotely on the WestGrid system, and we need to build into them some criteria that tells them when to st
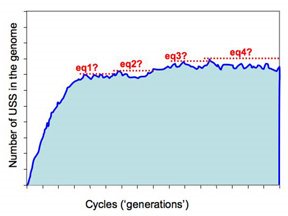 op. The present approach is illustrated in this figure.
op. The present approach is illustrated in this figure.The program checks for equilibrium by keeping track of the largest number of USS present in any previous cycle. If a sufficiently long time elapses without this number being exceeded, the run is treated as being at equilibrium and the long-range average number of USS calculated. So the dotted red lines in the figure illustrate four 'local maximum' USS numbers, and the time elapsed before that number was exceeded.
So how long is 'sufficiently long'? Because different settings affect whether it takes 1000 or 1,000,000 cycles to get to equilibrium, 'sufficiently long' is set as a percent of the total number of elapsed cycles. In the past I think I've used 20%, but I don't have any special reason to think this is the best choice. Beginning by doing lots of fast simulations should give us a much better understanding of what matters here.
















No comments:
Post a Comment
Markup Key:
- <b>bold</b> = bold
- <i>italic</i> = italic
- <a href="http://www.fieldofscience.com/">FoS</a> = FoS